|
Institut für Biologie III Schänzlestr. 7800 Freiburg Federal Republic of Germany Chromosomes are commonly regarded as conservative structures. in
which an exact amount of genetic information is arranged in a definite
sequential order This order is normally preserved when information
is exchanged between chromosomes. and guaranteed by a set of recombination
enzymes that function only with paired sectors of homologous DNA.
But processes such as inversion. deletion. duplication and translocation,
often involving recombination between apparently non-homologous
chromosomal regions, can alter this sequential order. Chromosomal
rearrangements resulting from such events have heen observeld in
some cases with a disturbingly high frequency thus inviting speculation
about their biological significance. The processes involved have
heen termed collectively "illegitimate recombination". reflecting
our bias for conventional pathways based on sequence homology, but
evidence is accumulating which may legitimatize them as important
aspects of evolution or even differentiation. The presence of IS
elements in the chromosome of E. coli was originally revealeld by
the transposition of these DNA elements form their natural positions
into indicator systems, resulting in a recognizable mutant phenotype.
If for exampler the gal operon is used as an indicator system. mutations
can be isolated which are caused by the integration ofan IS element
into one of the three structural genes of the operon. Not only is
the thus interrupted structural gene inactivated but the expression
of the promoter distal genes is also abolished These mutations therefore
are strongly polar. The analysis of the nature of such mutations
has bten facilitated by the isolation ofgal transduc ing phage and
the development of techniques for examining heterolduplex DNA in
the electron microscope. With help of these tools it is possible
to inspect hybrid DNA molecules consisting of one DNA strand carrying
the strongly polar mutation paired with the complementary strand
of l dgal in the electron microscope. The strongly polar mutation
is seen as a single stranded DNA loop emerging from a position in
the double stranded heteroduplex molecule which corresponds to the
map position of the mutation. Analysis of various independently
isolated strongly polar mutations with the above technique revealed
the existence of different categories of IS elements. The elements
were numbered according to the order of their detection. ISI is
about 800 nucleotide pairs Iong, while IS2, IS3. IS4 and IS5 are
each approximately 1400 basepairs long (For review see Starlinger
and Saedler, 1976.) In the following paragraphs we will concentrate
on the topics listed below. A. Chromosomal Rearrangements Mediated by IS1 IS1 a known to occur in multiple copies in the chromosome of E. coli K12 (Saedler and Heiss, 1973). They also seem to be integral parts of at least some bacterial plasmids like F+ and R (Hu et al., 1975). The formation of new chromosomal sequences can result from translocation, duplication, inversion or deletion of genetic material. All these events seem to playa role in the evolution of plasmids as well as chromosomes. ISelements appear to be responsible for some such chromosomal rearrangements. Non-adjacent chromosomal regions can be brought together by deletion of the intermittent genetic material. resulting in a new chromosomal order. This reaction has been studied extensively in ISl induced deletion formation (Reifand Saedler, 1975, 1977). The termini of the integrated ISI elements are most important in this process. ISl is retained in the deletion, thus allowing further rounds of rearrangements. IS1 can be considered as a generator for deletions. sometimes fusing the structural genes of the gal operon to other promoters and thus creating a new control circuit (Reifand Saedler, 1977). It is not yet clear. however, which enzymes are involved in this rather unusual type of recombination. Apparently the normal recombination pathways of E. coli are not involved. However, recently mutants were isolated which are deficient in ISl induced deletion formation. Such mutants may be helpful in the analysis of the enzymes involved in illegitimate recombinational events (Nevers and Saedler. 1977). B. IS Elements Are Also Found in Strategical Positions on Certain Plasmids The R-factors of the fi+ class are composed of two units. each capable of replicating autonomously if dissociated from each other. The RTF unit codes all functions necessary for cell to cell contact. thus allowing the transfer of the plasmid. The r-determinant carries most of the antibiotic resistance genes. An ISI-element separates the RTF unit from the r-determinant at each junction. Both ISI elements are oriented in the same direction (Hu et al., 1975: Ptashne and Cohen, 1975 ). This finding suggests a model to explain the formation and dissociation of R-factors as well as the amplification of the antibiotic resistance genes. Rownd and Mickel ( 1971) showed tha t R-factors can dissociate into the RTF and the r-determinant in Proteus mirabilis. Dissociation may occur by recom bination between the two homologues IS 1 substrates of the co-integrate plasmid generating two units, each containing an ISI. Fusion results from the reverse reaction. Amplification of antibiotic resistance genes could be due to recombination between the homologous ISl elements of different r-determinant molecules, leading to co-integrate plasmids containing multiple copies of the r-determinant units. In addition to ISI other IS-elements are also observed on R-factors, either as mutations or as integral parts of the molecule. For example in R6 of R 100-1 IS2 is found at a position within the transfer genes at which it does not cause a transfer defective mutation but rather contributes to the transfer positive character of the plasmids (Hu et al., 1975). Many of the antibiotic resistance genes can transpose to the various other DNA molecules (Cohen and Kopecko, 1976). At least one of the transposons is flanked by a known IS element (Mac Hattie and Jackowski, 1977). In the evolution of R-plasmids, IS-elements therefore seem to play an important role. C. Detection of Mini-Insertion DNA Elements The detection of IS elements using the heteroduplex technique is
limited by the size of the integrated ONA element. If for example,
an IS element is an order of magnitude smaller than ISl to IS5,
it cannot be readily recognized as a single stranded loop using
the heteroduplex technique. To analyse very small insertions another
technique is more adequate. If suitable restriction fragments are
available, one containing an integrated mini-insertion and the same
fragment without they will band at different molecular weight positions
when subjected to electrophoresis in agarose or polyacrylamide gels.
In this manner two mutants were shown to be due to the integration
ora very small picce of additional DNA. Fig. 1 gives the pattern
ora Hind II, Hind III double digest of various plasmid DNAs. Slot
3 shows the pattern of the parental plasmid pDG1, which is Gal positive.
The pattern of pDGI2, in which an IS2 is integrated in the control
region of the gal operon, eliminating expression of the gal genes,
is presented in slot 4. Note the appearance of the new bands (e
and f) and the shift in molecular weight of one band (from a to
b), due to the integration of IS2. Slots I and 5 show the pattern
of two independent Gal positive revertants obtained from plasmid
pDG 12. Note the increase in molecular weight of only band e in
both mutants. This can only be explained by assuming that a small
inscrtion is present in band e. Using appropriate markers (slot
2) as references, the increase in molecular weight can be calculated.
Mutation I (slot 5) is due to thc integration of about a 115 basepair
long piece of DNA, while the other mutation (slot I) is about a
60 basepair insertion. The former has been called IS6 and the latter
IS7. Both insertions confer a Gal positive phenotype to the cell
carrying the plasmid. Since they seem to have integrated into IS2,
they either destroy the polar signal on IS2 or, more likely, each
carries its own turn-on signal. Recently we sequenced both IS6 and
IS7 and compared their DNA sequence of IS2 in the region of integration
of these mini-insertions. It is quite obvious that both IS6 and
IS7 can be derived from IS2 sequences 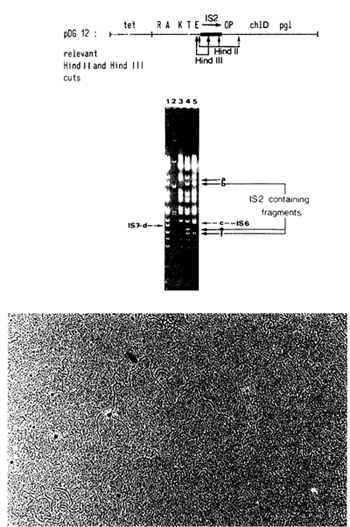
in a complicated manner. That is, genetic information from both DNA strands of IS2 seems to have multiplied and re-integrated in a rearranged fom, resulting in the formation of a turn-on signal. (For detailed discussion see Ghosal and Seadler, 1978 ) Conclusions IS elements are natural components of the E. coli chromosome. They can translocate from one position in the chromosome to another. Besides stimulating a number of illegitimate recombinational events, like deletion and transposition, of other gene, which is thought to be of evolutionary importance, they also carry signals necessary for gene expression (Saedler et al,.1974; Ghosal and Saedler, 1977, 1978). Similar events are also known to occur in higher organism (Nevers and Saedler, 1977). References Cohen, S. N., Kopecko, D. J.: Structural evolution of bacterial
plasmids: Role of translocating genetic elements and DNA sequence
insertions. Federation Proceedings 35, 2031-2036 (1976) |