|
1 BCDP, Program Resources Inc., NCI-Frederick Cancer
Research Facility, Frederick, MD 21701, USA
The techniques of molecular biology have had a dramatic effect on the advancement of human genetics. In particular, the development of restriction fragment length polymorphisms (RFLPs) has allowed researchers to generate genetic markers for virtually any region of the human genome. Most RFLPs occur when a mutation creates or deletes a recognition site for a restriction enzyme, generating a DNA fragment of altered size. In the simplest case this will create two alleles. A DNA probe which hybridizes to this fragment will detect the presence of these alleles in the DNA from different individuals. Probes used to detected RFLPs have been derived from both cloned genes and randomly isolated DNA segments. Thus, each RFLP is a genetically inherited marker for a precise location on a chromosome. By analyzing the inheritance of RFLPs in families, linkage analysis techniques can be used to determine the order and genetic distance between different polymorphic probes [1]. When this information is combined with data on the physicallocation of the probes, agenetic map of a chromosome can be constructed [2]. Genetic maps are powerful tools, useful in the detailed molecular analysis of biological problems. By studying the inheritance of polymorphisms in families afflicted with a genetic disease, the approximate chromosomal location of the gene responsible for the disease can be determined. Linkage with RFLP markers has been detected for several human disease genes including Huntington's disease [3], muscular dystrophy [4], and cystic fibrosis [5-7]. Linkage is often a critical step in the eventual isolation of the gene itself [8]. RFLPs have also provided critical data in the analysis of specific chromosomal abnormalities in human tumors. DNA markers can be used to distinguish between the two homologous chromosomes in the normal cells of a patient and, when compared with DNA from a tumor, can detect the loss of a specific chromosome or chromosomal region. Such analysis helped to identify the region containing the gene responsible for retinoblastoma [9] and regions containing putative genes involved in Wilm's tumor and renal cancer [10,11] among others. In this report we present data on several newly detected RFLPs in biologically important genes (Table 1), describe in detail the identification of new RFLPs in three loci, and discuss the potential usefulness of genetic polymorphisms in the analysis of human disease.
The c-raf:.1 gene is a member of the family of serine protein kinases and is the cellular homologue of the v-raf gene. vraf was identified as the transforming gene of the murine 3611 acute transforming retrovirus [12]. It is believed that part of the function of the c-raf-1 gene is to act as an intracellular signal transmission molecule for certain hormones or growth factors [13]. The human c-raf-1 gene has been localized to chromosome 3 at bands p24-25 [14]. Using cDNA probes from the human c-raf-1 gene, we have searched for the presence of RFLPs in this locus. The pTuc-8 probe detects a polymorphism with the enzyme EcoRI, with alleles of15 and 12 kb (Fig. 1). In an analysis of 71 unrelated Caucasians, we found a frequency of 0.87 for the 15-kb allele and 0.13 for the 12-kb allele (Table 1). In addition, the EcoRI poly Table 1. Newly described RFLPs
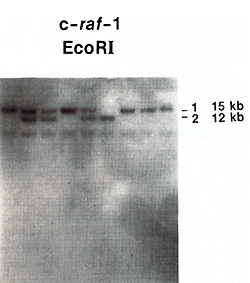 Fig.1. EcoRI RFLP in the c-raf-l gene. DNA from nine
individuals was hybridized with the pTUC-8 cDNA probe. morphism was shown to segregate properly in three large three-generation
pedigrees (data not shown). Specific deletions in the short arm
of chromosome 3 have been described in patients with small-cell
lung cancer (SCLC) and renal cell carcinoma [15,16]. U sing RFLPs
within the c-raf-110cus, we have examined DNA from a total of 83
human lung carcinomas (Sidthansen et al., unpublished). In an analysis
of ten paired (normal versus tumor) SCLC DNA samples, five informative
cases showed loss of heterozygosity at this locus. A BglI polymorphism
which shows 50% heterozygosity in a normal population was used to
analyze 73 unpaired lung carcinoma DNAs. Fifteen of 31 non-SCLC
samples showed heterozygosity; however, none of the 42 SCLC samples
were heterozygous. This striking apparent loss of heterozygosity
at the c-raf-1 locus in SCLC provides evidence that the c-raf-1
locus is deleted in smallcell lung carcinoma. Recently, Siezinger
et al. [17] used RFLPs in the c-raf-l gene to detect linkage between
this gene and the gene responsible for Von Hippel-Lindau syndrome,
an inherited cancer syndrome. The polymorphism described here can
be used to further study families with this disease in order to
pinpoint the location of the gene. We used probes from the interleukin-1alpha
(IL-1alfa) gene to screen for RFLPs at this locus. IL-1alfa is a
secreted protein that is involved in the stimulation of the growth
of lymphocytes. While secreted principally from macrophages, the
protein is also produced from keratinocytes and lymphocytes and
plays a role in the stimulation of lymphocytes and fibroblasts [18].
Using an IL-1alfa cDNA probe we detected a two-allele polymorphism
with the enzyme TaqI, and found that these alleles have frequencies
of 0.74 and 0.26 (Table 1 ). In addition, we found that a fragment
of this gene, detected by several different enzymes, showed a high
degree of variability between individuals. This type of variation
is characteristic of 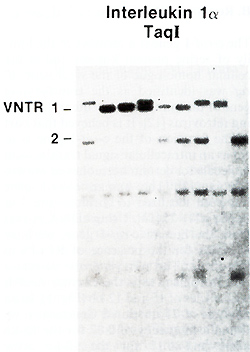
polymorphisms known as VNTRs (variable number of tandem repeats).
VNTRs are regions of DNA containing a group of tandem-repeated sequences
[19]. The number of repeats at a given locus is often highly variable
between different individuals, making these polymorphisms very informative
for genetic analyses. Examination of the published genomic sequence
of the genomic IL-l alfa gene [20] revealed the presence of a group
of tandem repeats in the intervening sequence between the fifth
and sixth exons (Fig. 2). The sequence of these repeats is typical
of those found in other VNTRs which have been characterized.. The
ILlalfa gene has been mapped to chromosome 2q13-21 and the polymorphisms
we have described will be useful genetic markers for this region.
In addition, these polymorphisms can be used to test whether the
IL-l alfa gene is linked to any human genetic disease for which
family pedigrees are available. 
Cystic fibrosis (CF) is the most common fatal genetic disease among Caucasians; it is caused by a recessive mutation at a single locus. The gene responsible for cystic fibrosis has been linked to polymorphic markers that map to chromosome 7q31 [21]. Recently, the location of the gene has been narrowed to a 700-kb region between the probes XV2c and pJ3.11 [22]. In order to clone and characterize additional sequences from this region, we have used chromosome jumping [23, 24] to isolate sequences surrounding the pJ3.11 locus. Briefly, chromosome jumping involves the circularization of large DNA fragments and the cloning of the ends of such fragments. By generating a library of chromosome-jumping clones and screening with a probe from the pJ3.11 locus, we have been able to isolate sequences spanning 300-400 kb surrounding pJ3.11 (Fig. 3). Using probes from this region we have searched for RFLPs and have discovered three new polymorphisms to date (Table 1). Two of these RFLPs are detected with the CF29 probe, which is located approximately 150 kb from pJ3.11, away from the CF gene. The third RFLP is detected with CF70-32, a jumping clone that lies closer to the CF gene (Fig. 3; unpublished data). While all three of these RFLPs are fairly informative, in preliminary analysis they all show a high degree of linkage disequilibrium with the MspI RFLP detected by pJ3.11. In other words, all four of these RFLPS reveal very little new information and increase the heterozygosity only from 53% to 58%.
Further progress in the molecular analysis of human genetic disease will rely on the development of increasingly detailed chromosome maps. At present, most of the maps of human chromosomes were constructed principally with anonymous DNA markers, not known to be part of genes. We have begun to characterize RFLPs in biologically important genes, many of which have been mapped to specific chromosomallocations by in situ hybridization. The list of genes in which we have detected RFLPs includes the proto-oncogenes and growth factors craf-1, c-jun, c-ovc-2, and IL-l alfa, and cell surface molecules and receptors such as the retinoic acid receptor, the T -cell receptor zeta gene, and the T 4 gene, receptor for HIV. In addition to providing markers for specific chromosomallocations, the RFLPs in these genes will allow them to be tested as candidate genes for human genetic diseases.
1. Botstein D, White R, Skolnick M, Davis RW (1980) Construction ofa genetic linkage map in man using restriction fragment length polymorphisms. Am J H um Genet 32:314-331 2. White R, Leppert M, Bishop DT, Barker D, Berkowitz J, Brown C, Callahan P , Holm T, Jerominski L (1985) Construction of linkage maps with DNA markers for human chromosomes. Nature 313:101-105 3. Gusella JV, Wexler, NS et al. (1983) A polymorphic DNA marker genetically linked to Huntington's disease. Nature 306:234 4. Davies KE, Pearson PL et al. (1983) Linkage analysis of two cloned DNA sequences flanking the Duchenne muscular dystrophy locus on the short arm of the human X chromosome. Nucleic Acids Res 11:2303-2312 5. Tsui L-C, Buchwald M, Barker D, Braman JC, Knowlton R et al. (1985) Cystic fibrosis locus defined by a genetically linked polymorphic DNA marker. Science 230: 1054-1057 6. Wainwright BJ, Scambler PJ, Schmidtke J, Watson EA, Law H- Y, Farrall M, Cooke HJ, Eiberg H, Williamson R (1985) Localization of cystic fibrosis locus to human chromosome 7cen-q22. Nature 318: 384-386 7. White R, Woodward S, Leppert M, O'Connell P, Hoff M, Herbst J, Lalouel J-M, Dean M, Vande Woude G (1985) A closely linked genetic marker for cystic fibrosis. Nature 318:382-384 8. Monaco AP, Neve RL, Colletti-Feener C, Bertelson CJ, Kurnit DM, Kunkel LM (1986) Isolation of candidate cDNA's for portions of the Duchenne muscular dystrophy gene. Nature 323:646-650 9. Cavenee WK, Dryja TP, Phillips RA, Benedict WF Godbout R, Gallie BL, . Murphree AL, Strong LC, White RL (1983) Expression of recessive alleles by chromosomal mechanisms in retinoblastoma. Nature 305: 779- 784 10. Koufos A, Hansen MF, Copeland NG, Jenkins NA, Lampkin BC, Cavenee WK (1986) Loss of heterozygosity in three embryonal tumors suggests a common pathogenetic mechanism. Nature 316: 330- 334 11. Zbar B, Brauch H, Talmadge C, Linehan M (1987) Loss of alleles of loci on the short arm of chromosome 3 in renal cell carcinoma. Nature 327: 721 12. Rapp UR, Goldsborough MD et al. (1983) Structure and biological activity of v-raf, a unique oncogene transduced by a retrovirus. Proc Natl Acad Sci USA 80:4218 13. Rapp UR, Storm SM, Cleveland JL (1987) Oncogenes: Clinical Relevance. Hematology 31 :450-459 14. Bonner T, O'Brien S1, Nash WG, Rapp UR (1984) The human homologs of the raf (mil) oncogene are located on human chromosomes 3 and 4. Science 223: 71- 74 15. Whang-Peng 1, Kao-Shan CS, Lee EC (1982) Specific chromosome defect associated with human small-ccll lung cancer: deletion 3p(14-23). Science 215:181-182 16. Kok K, Osinga 1, Carritt B, Davis MB, van der Hout AH, van der Veen AY, Landsvater RM et al. (1987) Deletion ofa DNA sequence at the chromosomal region 3p21 in all major types of lung canccr. Nature 330: 578-581 17. Seizinger BR, Roleau GA et al. (1988) Yon Hippel-Lindau disease maps to the region of chromosome 3 associated with renal cell carcinoma. Nature 332: 268 -269 18. Oppenheim 11, Kovacs E, Matsushima K, Durum SK ( 1986) There is more than one interleukin 1. Immunol Today 2: 45- 56 19. Nakamura Y, Leppert M, O'Connell P, Wolfe R, Holm T, Culver M, Martin C, Fujimoto E, Hoff M, Kumlin E, White R (1987) Variable number of tandem repeat (YNTR) markers for human gene mapping. Science 235: 1616-1622 20. Furutani Y, Notake M, Fukui T, Ohue M, Normura H, Yamada M, Nakamura S (1986) Complete nucleotide sequence of the gene for human interleukin 1 alpha. Nucl Acids Res 14: 3167 -3179 21. Dean M (1988) Molecular and genetic analysis of cystic fibrosis. Genomics 3:93-99 22. Drumm ML, Smith CL, Dean M (1988) Physical mapping of the cystic fibrosis region by pulsed-field gel electrophoresis. Genomics 2: 346-354 23. Collins FS, Weissman SM (1984) Directional cloning of DNA fragments at a large distance from an initial probe: A circularization method. Proc Natl Acad Sci USA 81:6812-6816 24. Collins FS, Drumm ML, Cole 1L, Lockwood WK, Vande Woude FG, Iannuzzi M C ( 1987) Construction of a general human chromosome jumping library, with application to cystic fibrosis. Science 235: 1046-1049 |